code
- 📰 RSS: I’ve swapped out Feedbin for iCloud as the backend for NetNewsWire. Although I really like the Feedbin service, my primary use of it is to access my feeds via a web browser at work. In an attempt to limit my feed reading to just once in the morning and once in the evening, switching to iCloud means that only my personal iPhone has access.
- 👓 Read It Later: Switching to Safari’s Reading List for this. The feature is well integrated into the system and more than sufficient for my needs.
- 🎧 Podcasts: After some back and forth and back again, over to Apple Podcasts.
- As should be apparent from above, creating useful Shortcuts for Apple Notes is straightforward. In some sense, it is the Shortcuts that I’m finding really useful. Apple Notes is just the final destination for the content.
- I’ve set up widgets by focus mode so that the most recent notes are shown on my Home Screen in the right context. These are restricted to a particular folder and sorted by date modified.
- The formatting options are comprehensive, including table support.
- I think I like the feature where checking off tasks moves them to the bottom of the list. Most of the time, this is what I want.
- The iCloud web app is convenient for using notes on my Windows work PC. Unfortunately, I’ve found the syncing to be rather unreliable here, where notes just don’t show up in the web app sometimes.
- I don’t share notes as often as I expected. When I do, it works really well.
- Not really specific to Apple Notes, but I stole an idea from Matthew Cassinelli to aggregate all of these Notes Shortcuts into one super Shortcut that creates a list of Shortcuts to choose from.
- Searching is too limited. In particular, you can’t narrow searches to particular folders. Most of the time I either only want to search my meeting notes or not include them. I had to set up a Shortcut that takes a search term as an input and then asks me to specify which folder to search. This should be built into the app’s search field.
- Linking among notes isn’t really supported. You can sort of do this with url searches for note titles. Pretty clunky though.
- Given how much I use Shortcuts for Apple Notes, it is frustrating how little you can do when creating a note. In particular, you can’t style text or add tags. Every time I use Shortcuts to create a note, the first thing I have to do is apply title and heading styles and convert any words prefixed with a # into an actual tag.
- Blog posts
- Drafting long emails
- Capturing and processing transitory texts, which Drafts is really optimized for
- One less account to worry about. Not that it was a big deal, but now I don’t need to know the various setup details for my personal email. Once I’ve logged into my iCloud account, my email is ready.
- I appreciate Apple’s commitment to privacy and trust that they’ll apply this commitment to my email account.
- I’m already paying for iCloud+ and, so, might as well use this feature and save some money by not paying for separate email hosting.
- I’m actively using Reminders and Notes in iCloud.com and the Mail interface there is decent, certainly better than the rudimentary one offered by my previous email host.
- Document syncing is far more reliable. This isn’t entirely Agenda’s fault. I’m restricted by corporate policy from using iCloud Documents, so have been using Dropbox sync for Agenda. I often have to wait an indeterminate, though long, time before documents sync across my devices. Craft sync has been instantaneous and very reliable.
- Having access to my documents from a web browser is great. I’ll be back to working from the office on a Windows laptop soon and won’t have access to my iPad. So, web access will be important.
- Performance is much better on Craft. Agenda often freezes in the middle of typing and suffers from random crashes. This could very well be something about my particular setup, though it doesn’t happen in any other apps.
- 📚 for Apple Books
- 💵 for The Economist
- 📰 for NetNewsWire
- 👓 for Safari Reading List
Even more defaults
As a follow up to my Duel of the Defaults post, I’ve made a few changes. These are all based on further adopting app defaults to simplify things.
These choices are largely motivated by an attempt to limit the number of inputs and potential for distractions. That said, these default apps are still powerful and effective.
Switching podcast apps, again 🎧
As predicted, after a couple of months with the Apple Podcasts app, I’m back to Overcast.
I think that Apple’s Podcasts app is great for anyone new to podcasts, given it has a strong focus on discovering new shows. I’m looking for a podcast app that simply plays my carefully curated, short list of podcasts. With Apple Podcasts, I kept finding new episodes of shows I didn’t intend to subscribe to in my queue.
Adding in the nice audio features in Overcast that boosts voices and trims silences, makes Overcast the right app for me.
Of course, having just made this decision and rebuilt my Overcast subscriptions, I see that Apple has now integrated Apple Music into their Podcasts app. Better ways of managing radio episodes in Apple Music is on my list of features I’d like to see. So, looks like I’m not actually settled on a podcasts app yet, which was a silly hope anyway.
Simplifying my personal iPhone
Now that I’ve separated my work and personal iPhones, I’m taking some time to simplify my personal device.
The biggest change is that for work, I’m now fully into the Office 365 product. So, email in Outlook, tasks in To Do, and notes in OneNote. Although I really liked using MindNode as my project and task manager and Apple Notes for my notes, I have to admit that this just works so much better for work tasks and with my office Windows PC. This has significantly reduced the demands on my personal phone, which was part of the point.
On the personal side, I’m sticking with Apple Notes and Reminders. These apps have seen lots of improvements recently and are more than sufficient for my needs. I’ve found that sharing notes with my family has been really useful, both as a way to work through family projects and to share reference material.
For music, I’m back to the Apple Music app. I liked the concept and design of the Albums app. I just found that I rarely used it. I am keeping MusicBox as the place to track albums I want to listen to (stuff gets lost in the Recently Added section of the Apple Music app). I’m also tagging albums there for particular moods, since I never seem to be able to remember relevant albums in the moment. MusicBox is a really nice, lightweight companion to Apple Music for these use cases.
For podcasts, I’ve switched back to Apple Podcasts from Overcast. This is really just an experiment with standardizing on stock Apple apps. I don’t expect it to last. Overcast has so many nice refinements and now also has better access to OS integrations that used to favour the Apple Podcasts app.
A couple of things that haven’t changed are getting news via Feedbin in NetNewsWire and journaling in DayOne. Although I’m intrigued by the new Journal app from Apple and will certainly test it out, I expect that I’ll keep my DayOne subscription and the close to 9,000 entries I’ve added.
Knowing my predilections for fiddling with my setup, I’ve intentionally kept my homescreen stable for a couple of years now. No doubt there will be further tweaks once iOS 17 is released. After that, I look forward to another stable setup that I use when necessary and otherwise leave alone.
☎️ 😱 Living dangerously for seven years with a corporate phone
For seven years now, I’ve been living dangerously by only using my corporate phone for everything. I knew this was wrong, yet couldn’t resist, until this week.
There were only two, day-to-day negative impacts of relying on a corporate phone.
The first, admittedly minor, though surprisingly annoying, one is that any explicit songs in Apple Music were blocked. It isn’t that I feel compelled to listen to explicit lyrics. Rather, there are lots of good songs with a few swear words thrown in, especially for the more high-energy rock I prefer for workouts. Sometimes clean versions are available, though they lack the power of the real versions.
The second, more systematic, one is that iCloud Drive support was blocked. This disabled some important features of many of my favourite apps, like Drafts, Mindnode, Soulver, MusicBox, and Albums. These apps became little islands of inaccessible data that didn’t share with my iPad or Mac, limiting their utility.
Of course the real reason this was a bad idea is exactly what IT says: you shouldn’t mix work with personal. IT has been (appropriately) locking down more and more of the phone, while also adding in VPN and other monitoring apps.
So, why did I do it? Two main reasons: I really don’t like carrying extra stuff and I saved the monthly cost of a personal data plan (given Canada’s rates, this is bigger than you might think).
There was no epiphany that led me to finally get a personal phone. Just a steady realization that meant it was time.
I picked up an iPhone 13 mini. I have no need for the latest phone and really appreciate the smaller size of the mini.
I opted for a Freedom Mobile plan and added in the Apple Watch plan (something I couldn’t do with the corporate phone). My biggest surprise so far is how nice it is to have the Apple Watch Ultra connected via cellular and leave the phone behind. The Apple Watch really is quite functional for my needs without the phone.
Better late than never to this. I’m glad to finally have made the right choice and am enjoying better partitioning of work from the rest of my activities.
I’m letting my Pinboard subscription expire. I’ve added 3,500 bookmarks since 2010 and the service was valuable when actively engaged in research. I haven’t looked up a bookmark in the past few years though. Now I’ll rely on Apple Notes and Micro.blog Bookmarks
Time to stop tracking my personal life ⏰
Through 2020, I built up an ornate system for tracking my time for both work and personal projects (like this one for reading). For most of 2021, I found this tracking really helpful.
I need to track my hours at work anyway, so using Timery and Shortcuts to automate much of this has been great. Having a strong sense of how long things take and ensuring good balance across projects are all benefits of time tracking.
For personal projects, though, I’ve been starting to feel a bit stressed by having a timer always running whenever I’m doing something, almost like I’m always in a race. At first, knowing how much time I was spending on particular things was great for my Year of the Tangible intention. This is well established now, and I haven’t been using the time reports for any personal projects. So, why am I creating anxiety for no benefit?
I’ve turned off all of my time tracking automations for personal projects. Despite some annoying bugs, ScreenTime is a good-enough replacement for keeping an eye on time spent on things like YouTube and social networking. A nice side benefit is that this also reduces the number of Shortcuts and other automation that I need to manage, allowing me to just enjoy my personal time.
Of course, I’ll keep tracking work projects, since the benefits far outweigh the costs there.
As an update to my earlier post about using MindNode for task management, I’ve refreshed my areas of focus and projects for work. I still find MindNode really helpful for this, especially for seeing the balance of projects across the areas of focus. In this case, I can see that I have many Process Improvement projects, which makes sense, given my company has a big push on Lean at the moment.
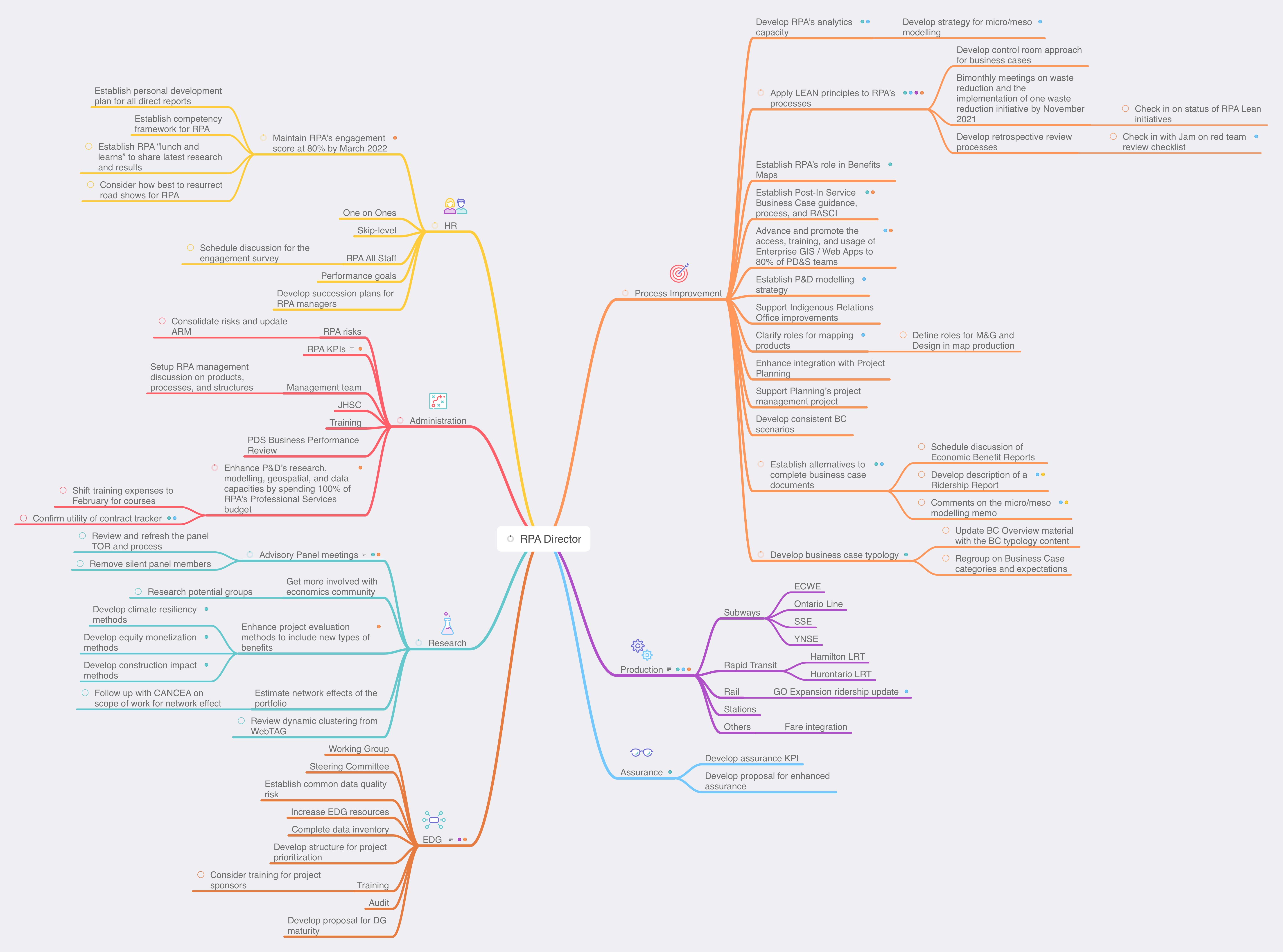
Now I can fill in next actions for each project and then sync with Reminders.
Lessons from using Apple Notes for three months
Back in September, I committed to using Apple Notes for three months. The goal was to focus on my use cases for writing, rather than fiddling with new apps continuously.
Here’s what I’ve identified so far. Many of the approaches and features that I’m using in these use cases are readily available in other apps and often Notes is not the most efficient choice. Now that I’ve documented these use cases, I’d like to use them to assess alternative apps.
Meeting notes
Thanks to Timery, I know that 60% of my working time is spent in meeting. So many meetings!
For each one, I create a note to capture ideas, useful information, and tasks. I’ve automated this with a couple of Shortcuts. The one I use the most is “Start My Next Meeting”. This presents me with a list of upcoming meetings. I choose from the list and it creates a meeting note, starts a Timery timer, and opens the link to start the video call (typically Teams). The meeting note has the name of the meeting as the title, adds tags for #meeting and the Timery project, adds the date and time of the meeting, a list of attendees, and any notes from the calendar event. From this structure, I can then add notes throughout the meeting and extract any tasks into Reminders later.
I used Agenda for these sorts of notes before, which was powerful.
Daily summaries
Occasionally, I find myself at the end of a week with no clear sense of what I actually accomplished. To help with this, for the past year I’ve been recording the top three things I’ve done in a day into Day One (the 5 Minute PM template has been great for this).
To augment this, I’ve been using another Shortcut to create a Daily Work Report. This makes a note of the meetings I attended, tasks I completed, and tasks I created. These all get saved to a Daily Notes folder. I then use the day’s work report to pull out the highlights for Day One. There’s some redundancy here, though I find the process of choosing just three things for Day One is helpful.
Overall, I think that Day One is a better app for this use case.
Project notes
For each of my projects, I create a project note that states the purpose or objective of the project, key stakeholders, and timelines. Then I accumulate relevant notes and documents while making progress on the project. Creating these are also done via a simple Shortcut. I’ve experimented with using checklists for tasks in these notes, but find it isn’t as effective as my approach with MindNode and Reminders.
Once I finish a project, the associated note gets cleaned up and moved to an Archive folder to keep it out of the way.
Research
This is a rather broad category and, unlike the previous use cases, is for both work and personal notes. Much of this is capturing facts, quotes, and sources. If it is project specific, they go to the project note. Some are more generic and are kept as a standalone note. All of them get tags to help provide some structure. This is where Apple Notes ability to accept almost anything from the share sheet is powerful.
The new Quick Notes feature has been interesting for research. The ability to quickly highlight and then resurrect content on websites is great. I find actually working with the quick notes is pretty clumsy though. They have to stay in the Quick Notes folder and choosing which one to send content to can be tricky. I think there’s some great potential here and will keep experimenting.
For any webpages that I want to archive, I use another Shortcut that creates a plain-text note of the webpage along with some metadata and then adds the link to Pinboard. This has been surprisingly useful for recipes, when all I really want are the ingredients and steps, rather than the long history of the recipe’s development.
Other nice features
In addition to these use cases, there are a few nice features of Apple Notes that are worth mentioning.
Challenges
There are a few things that don’t work as well as they should:
Other use cases
I’ve found a few use cases that don’t yet fit in with Apple Notes. I’m using Drafts for all of these:
Restricting myself to Apple Notes was a helpful trick for crystallizing my use cases. Now that I’m three months in, I think I’ll stick with Apple Notes for a while longer. I’ve built up a good ecosystem of Shortcuts for working with the app and, of course, now have lots of content in the app.
Switching to iCloud+ Custom Email Domain 📧
I’ve switched my personal email over to Apple’s custom email domain with iCloud Mail. A roughly ranked list of reasons for the switch is:
Setup was straightforward with clear instructions. Having said that, the only issue I had was that initiating the setup process simply didn’t work for a few weeks. I tried a couple of times a week and each time I just got a generic error. Then, for no apparent reason, one day it worked. I suspect this was just an issue with rolling out a new service.
I should point out that my email needs are very basic for this personal account. I don’t need many automated rules, tagging, or filtering. So, iCloud Mail is fine. I wouldn’t switch over my work account (even if corporate IT would allow it). I get something like 100x the email at work and need more sophisticated tools.
Besides the initial trouble with initiating the setup, everything has been working well for the past week. I’m well aware of Apple’s well-earned reputation for challenges with internet services and will be staying vigilant for at least the next few weeks. One of the great benefits of having my own domain name is the ease with which I can switch mail hosts.
Switching from Agenda to Craft, for now 📅🗒
In my corner of the internet, there’s a well trodden, twisted path of searching for the one true notes app. I’ve reached a fork in the path between Agenda and Craft. As I wrote earlier, I’ve been using Agenda for a while now and its date-based approach really suits my meeting-dominated work. Now, though, Craft has added calendar integration and I’m testing it out.
There are several things I really like about Craft, relative to Agenda:
On the downside, I do miss Agenda’s simplicity. Craft has lots of ways to organize notes (such as cards and subpages). Of course, you can mostly ignore this, but I like Agenda’s well-thought-through approach that didn’t require much deliberation about where to put things.
Of course, having just made this switch, Apple announced Quick Notes and I may well be back on Apple Notes in a few months.
Scheduling random meetings with a Shortcut ⚙️🗓
Staying in touch with my team is important. So, I schedule a skip-level meeting with someone on the team each week. These informal conversations are great for getting to know everyone, finding out about new ideas, and learning about recent achievements.
Getting these organized across a couple of dozen people is logistically challenging and I’ve developed a Shortcut to automate most of the process.
Borrowing from Scotty Jackson, I have a base in AirTable with a record for each team member. I use this to store all sorts of useful information about everyone, including when we last had a skip-level meeting. The Shortcut uses this field to pull out team members that I haven’t met with in the past four months and then randomizes the list of names. Then it passes each name over to Fantastical while also incrementing the date by a week. The end result is a recurring set of weekly meetings, randomized across team members.
The hardest part of the Shortcut development was figuring out how to get the names in a random order. A big thank you to sylumer in the Automators forum for pointing out that the Files action can randomly sort any list, not just lists of files.
I’m not sharing the Shortcut here, since the implementation is very specific to my needs. Rather, I’m sharing some of the thinking behind the code, since I think that it demonstrates the general utility of something like Shortcuts for managing routine tasks with just a small amount of upfront effort.
MindNode is the best mind mapping app for iOS
Continuing my plan to update App Store reviews for my favourite apps, up next is MindNode.
MindNode is indispensable to my workflow. My main use for it is in tracking all of my projects and tasks, supported by MindNode’s Reminders integration. I can see all of my projects, grouped by areas of focus, simultaneously which is great for weekly reviews and for prioritizing my work.
I’ve also found it really helpful for sketching out project plans. I can get ideas out of my head easily with quick entry and then drag and drop nodes to explore connections. Seeing connections among items and rearranging them really brings out the critical elements.
MindNode’s design is fantastic and the app makes it really easy to apply styles across nodes. The relatively recent addition of tags has been great too. Overall, one of my most used apps.
Trying out a new iPhone Home Screen 📱
With the release of iOS 7, I’m reconsidering my earlier approach to the Home Screen. So far I’m trying out a fully automated first screen that uses the Smart Stack, Siri Suggestions, and Shortcut widgets. These are all automatically populated, based on anticipated use and have been quite prescient.
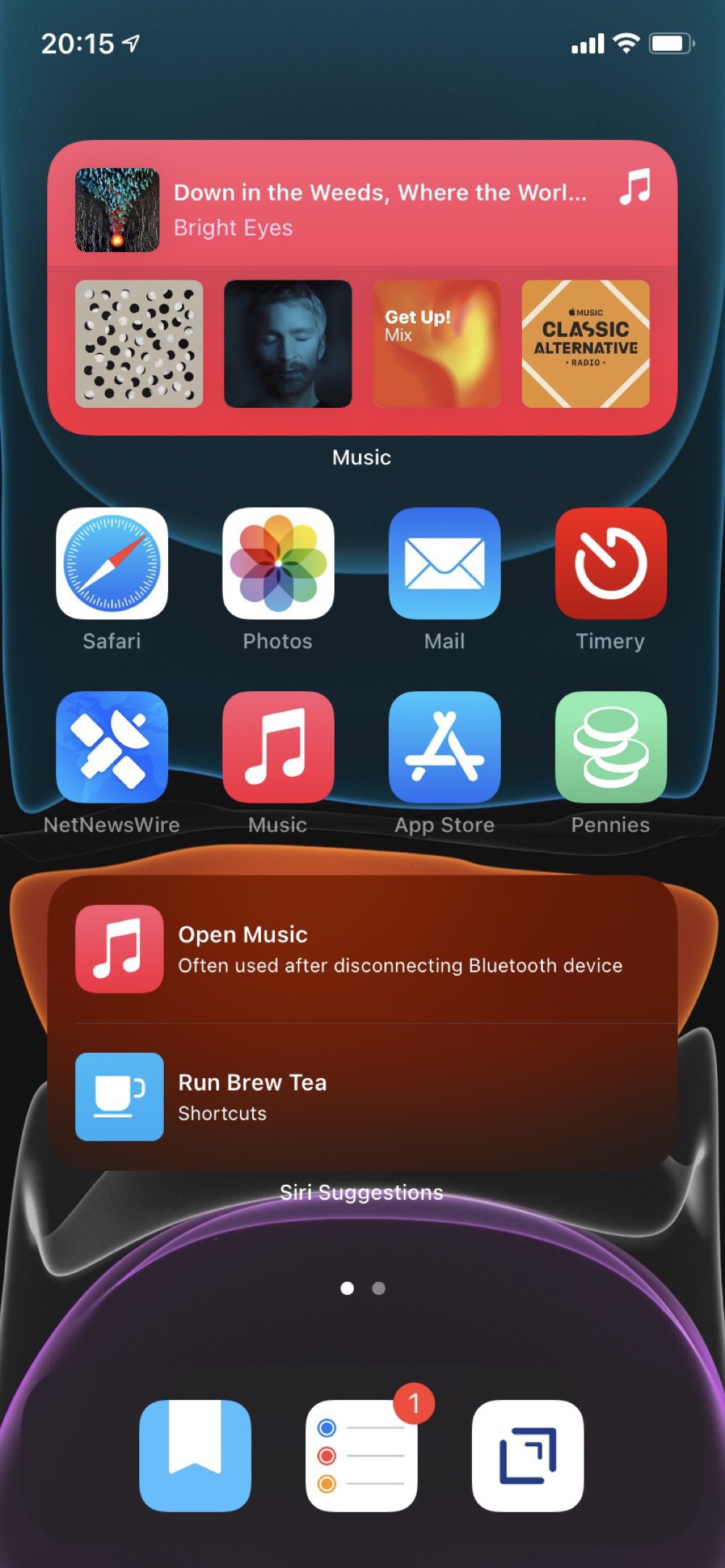
My second screen is all widgets with views from apps that I want to have always available. Although the dynamic content on the first screen has been really good, I do want some certainty about accessing specific content. This second screen replaces how I was using the Today View. I’m not really sure what to do with that feature anymore.
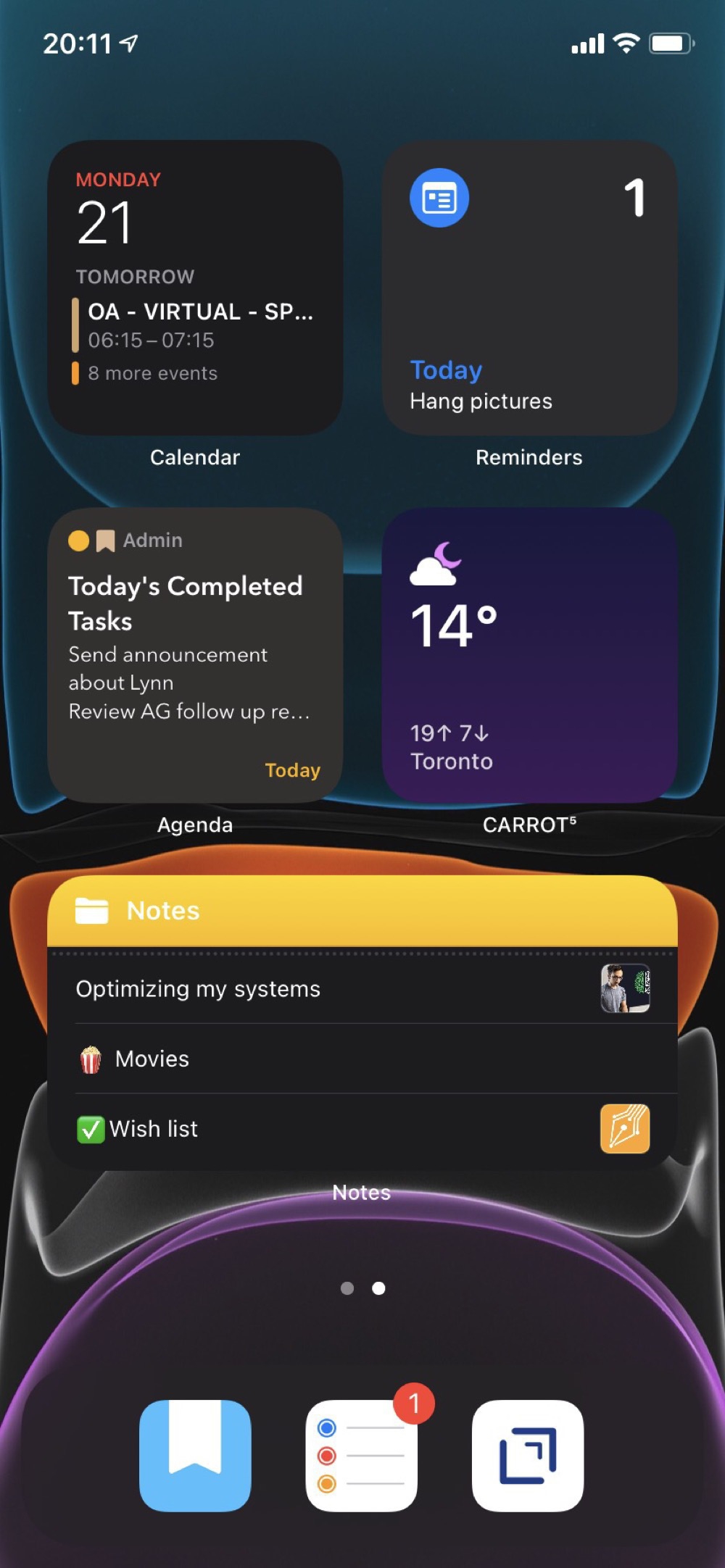
I’ve hidden all of the other screens and rely on the App Library and search to find anything else.
I still like the simplicity behind my earlier approach to the Home Screen. We’ll see if that is just what I’m used to. This new approach is worth testing out for at least a few weeks.
Reading Shortcut for the iPad 👓⚙️
I haven’t yet adopted the minimalist style of my iPhone for my iPad. Rather, I’ve found that setting up “task oriented” Shortcuts on my home screen is a good alternative to arranging lots of app icons.
The one I use the most is a “Reading” Shortcut, since this is my dominant use of the iPad. Nothing particularly fancy. Just a list of potential reading sources and each one starts up a Timery timer, since I like to track how much time I’m reading.
Here’s a screenshot of the first few actions:
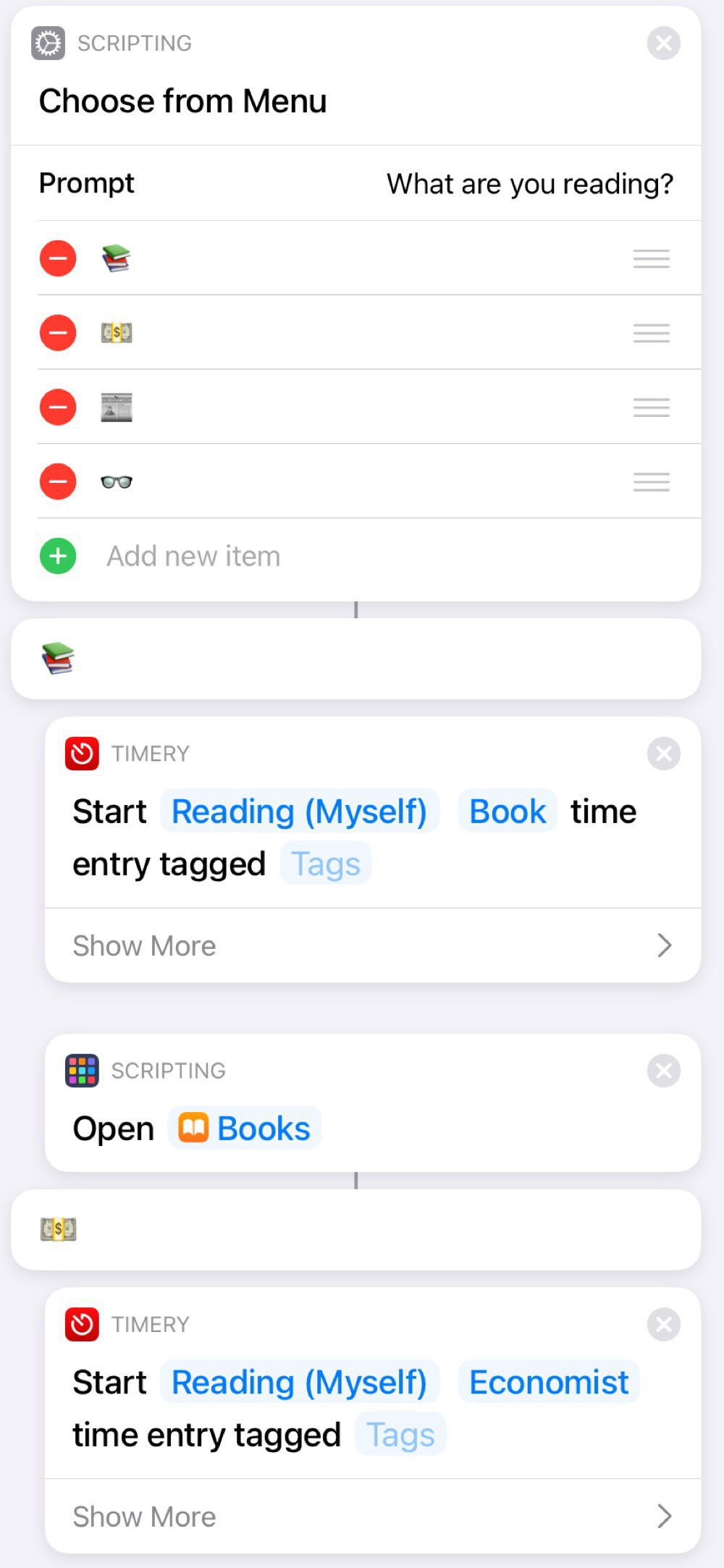
I like the bit of whimsy from using emoticons:
A nice feature of using a Shortcut for this is that I can add other actions, such as turning on Do Not Disturb or starting a specific playlist. I can also add and subtract reading sources over time, depending on my current habits. For example, the first one was Libby for a while, since I was reading lots of library books.
This is another example of how relatively simple Shortcuts can really help optimize how you use your iOS devices.
Reflection journal in Day One with an Agenda assist
I’ve been keeping a “director’s commentary” of my experiences in Day One since August 2, 2012 (5,882 entries and counting). I’ve found this incredibly helpful and really enjoy the “On This Day” feature that shows all of my past entries on a particular day.
For the past few months, I’ve added in a routine based on the “5 minute PM” template which prompts me to add three things that happened that day and one thing I could have done to make the day better. This is a great point of reflection and will build up a nice library of what I’ve been doing over time.
My days seem like such a whirlwind sometimes that I actually have trouble remembering what I did that day. So, my new habit is to scroll through my Today view in Agenda. This shows me all of my notes from the day’s meetings. I’ve also created a Shortcut that creates a new note in Agenda with all of my completed tasks from Reminders. This is a useful reminder of any non-meeting based things I’ve done (not everything is a meeting, yet).
I’m finding this new routine to be a very helpful part of my daily shutdown routine: I often identify the most important thing to do tomorrow by reviewing what I did today. And starting tomorrow off with my top priority already identified really helps get the day going quickly.
Different watch faces for work and home
watchOS 7 has some interesting new features for enhancing and sharing watch faces. After an initial explosion of developing many special purpose watch faces, I’ve settled on two: one for work and another for home.
Both watch faces use the Modular design with the date on the top left, time on the top right, and Messages on the bottom right. I like keeping the faces mostly the same for consistency and muscle memory.
My work watch face than adds the Fantastical complication right in the centre, since I often need to know which meeting I’m about to be late for. Reminders is on the bottom left and Mail in the bottom centre. I have this face set to white to not cause too much distraction.

My home watch face swaps in Now Playing in the centre, since I’m often listening to music or podcasts. And I have Activity in the bottom centre. This face is in orange, mostly to distinguish it from the work watch face.
Surprisingly, I’ve found this distinction between a work and home watch face even more important in quarantine. Switching from one face to another really helps enforce the transition between work and non-work when everything is all done at home.
The watch face that I’d really like to use is the Siri watch face. This one is supposed to intelligently expose information based on my habits. Sounds great, but almost never actually works.
I'm not analyzing COVID data, though I'm impressed with Ontario's open data
I’m neither an epidemiologist nor a medical doctor. So, no one wants to see my amateur disease modelling.
That said, I’ve complained in the past about Ontario’s open data practices. So, I was very impressed with the usefulness of the data the Province is providing for COVID: a straightforward csv file that is regularly updated from a stable URL.
Using the data is easy. Here’s an example of creating a table of daily counts and cumulative totals:
data_source <- "[data.ontario.ca/dataset/f...](https://data.ontario.ca/dataset/f4112442-bdc8-45d2-be3c-12efae72fb27/resource/455fd63b-603d-4608-8216-7d8647f43350/download/conposcovidloc.csv)"
covid_data <- read_csv(data_source) %>%
select(ACCURATE_EPISODE_DATE) %>%
rename(date = ACCURATE_EPISODE_DATE) %>%
group_by(date) %>%
summarise(daily_count = n()) %>%
mutate(cumulative_count = cumsum(daily_count))
From there we can make some simple plots to get a sense of how the case load is changing.
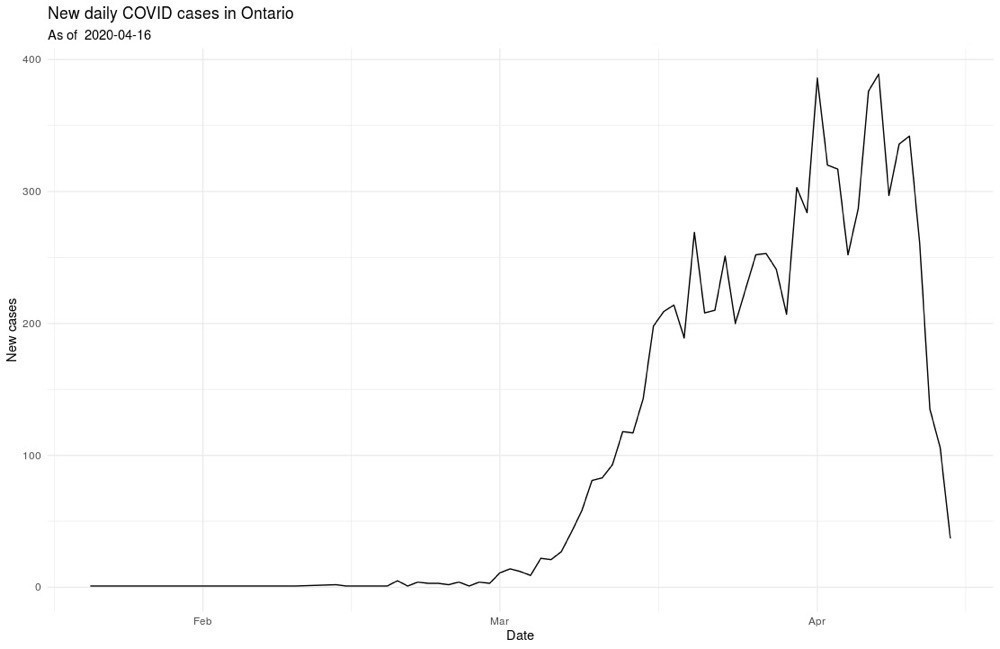
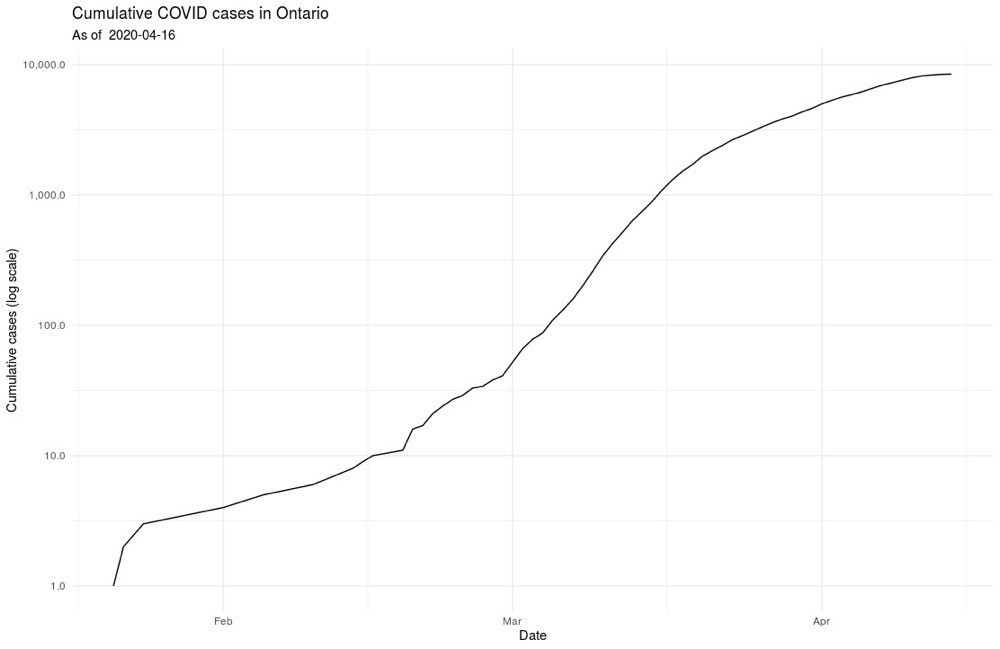
And, I’ll leave it at that, at least for public posting 🤓
Simple brew tea shortcut
Since I’m mostly stuck inside these days, I find I’m drinking more tea than usual. So, as a modification of my brew coffee shortcut, I’ve created a brew tea shortcut.
This one is slightly more complicated, since I want to do different things depending on if the tea is caffeinated or not.
We start by making this choice:
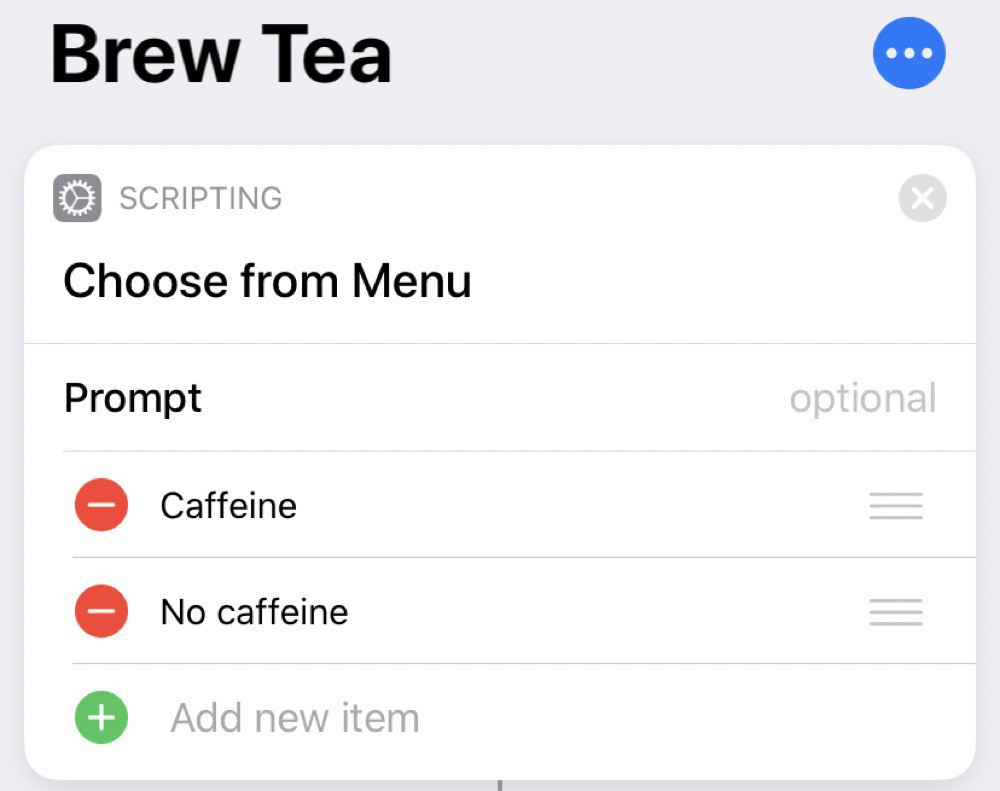
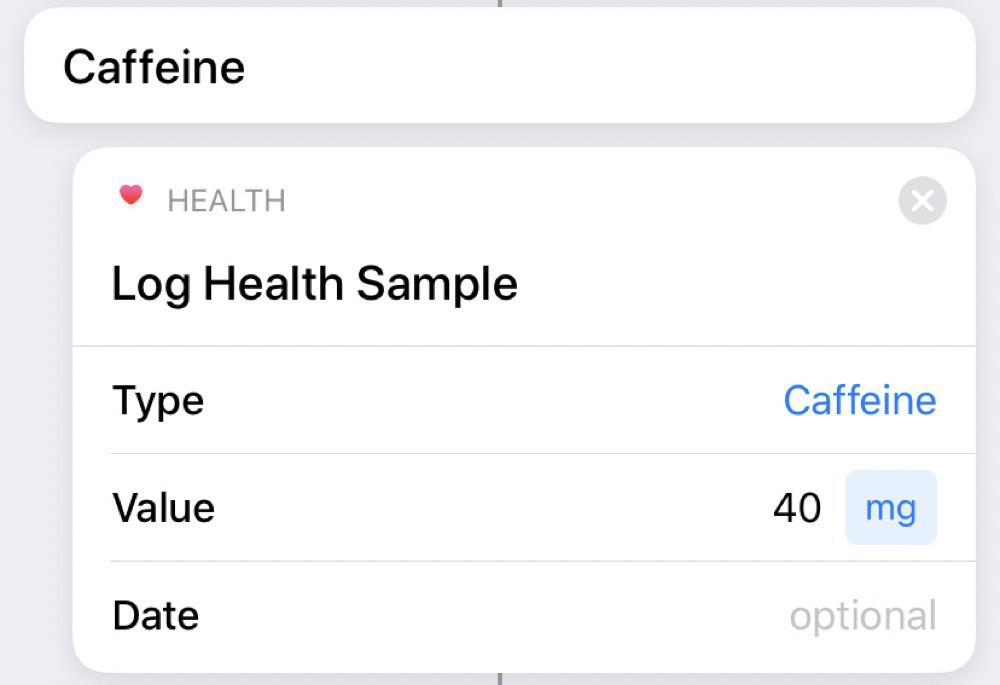
Then, if we choose caffeine, we log this to the Health app:
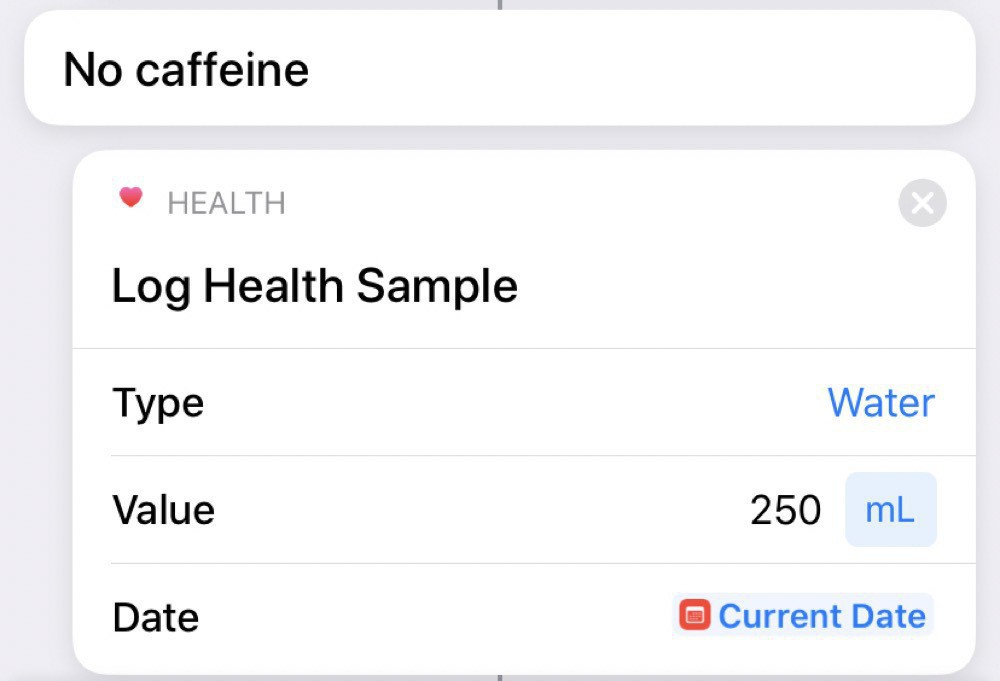
Uncaffeinated tea counts as water (at least for me):
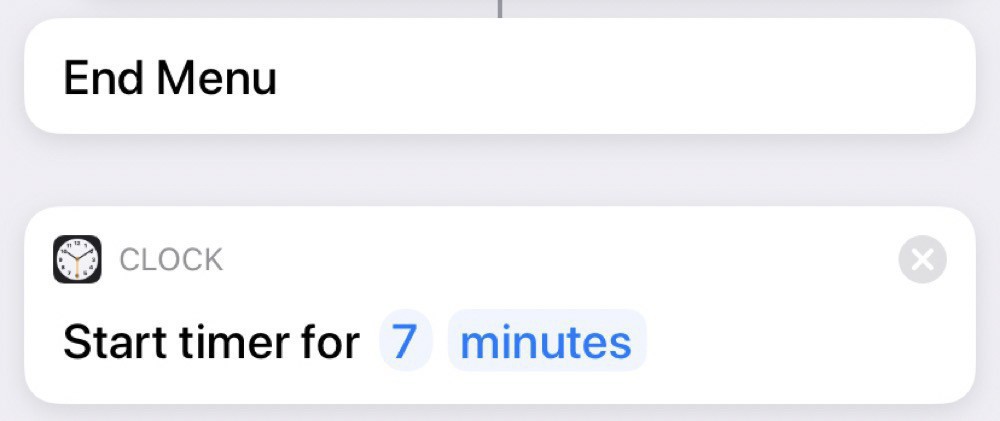
And, then, regardless of the type of tea, we set a timer for 7 minutes:
Running this one requires more interactions with Siri, since she’ll ask which type we want. We can either reply by voice or by pressing the option we want on the screen.
A simple Shortcut for tracking workout time
I’ve been tracking my time at work for a while now, with the help of Toggl and Timery. Now that I’m working from home, work and home life are blending together, making it even more useful to track what I’m doing.
Physical exercise is essential to my sanity. So, I wanted to integrate my Apple Watch workouts into my time tracking. I thought I’d be able to leverage integration with the Health app through Shortcuts to add in workout times. Turns out you can’t access this kind of information and I had to take a more indirect route using the Automation features in Shortcuts.
I’ve setup two automations: one for when I start an Apple Watch workout and the other for when I stop the workout:
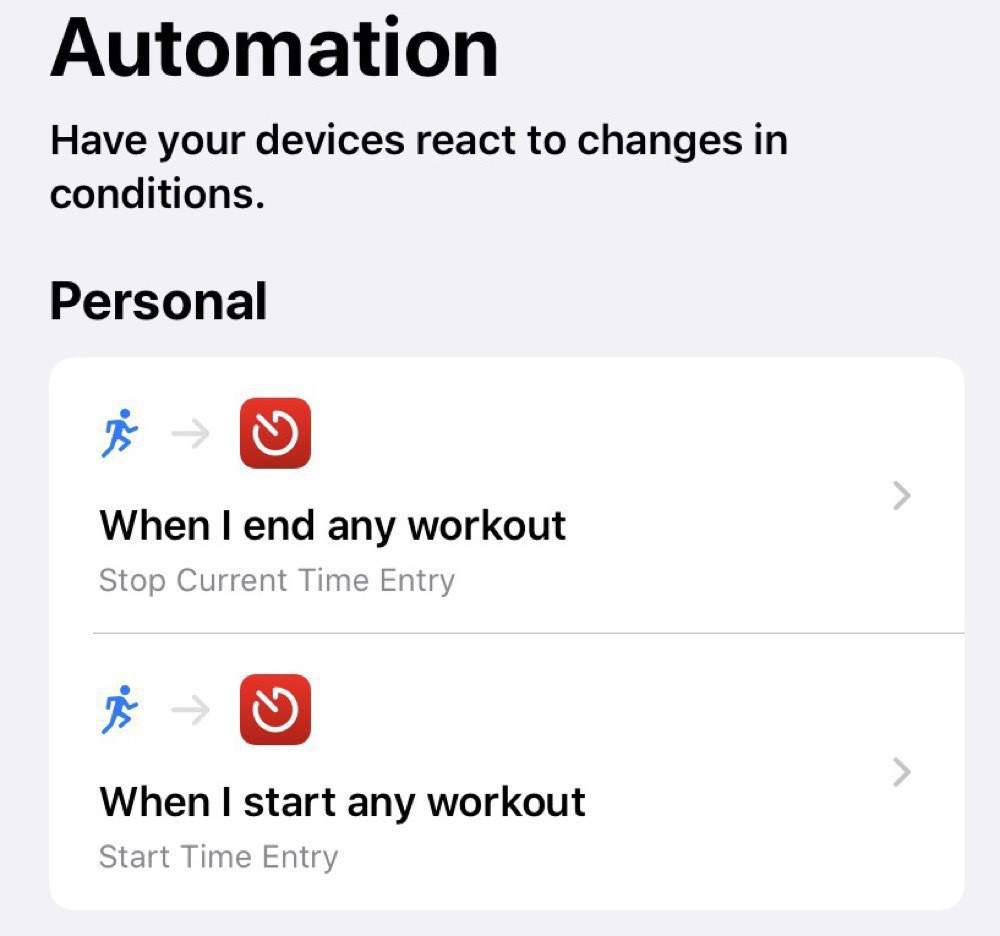
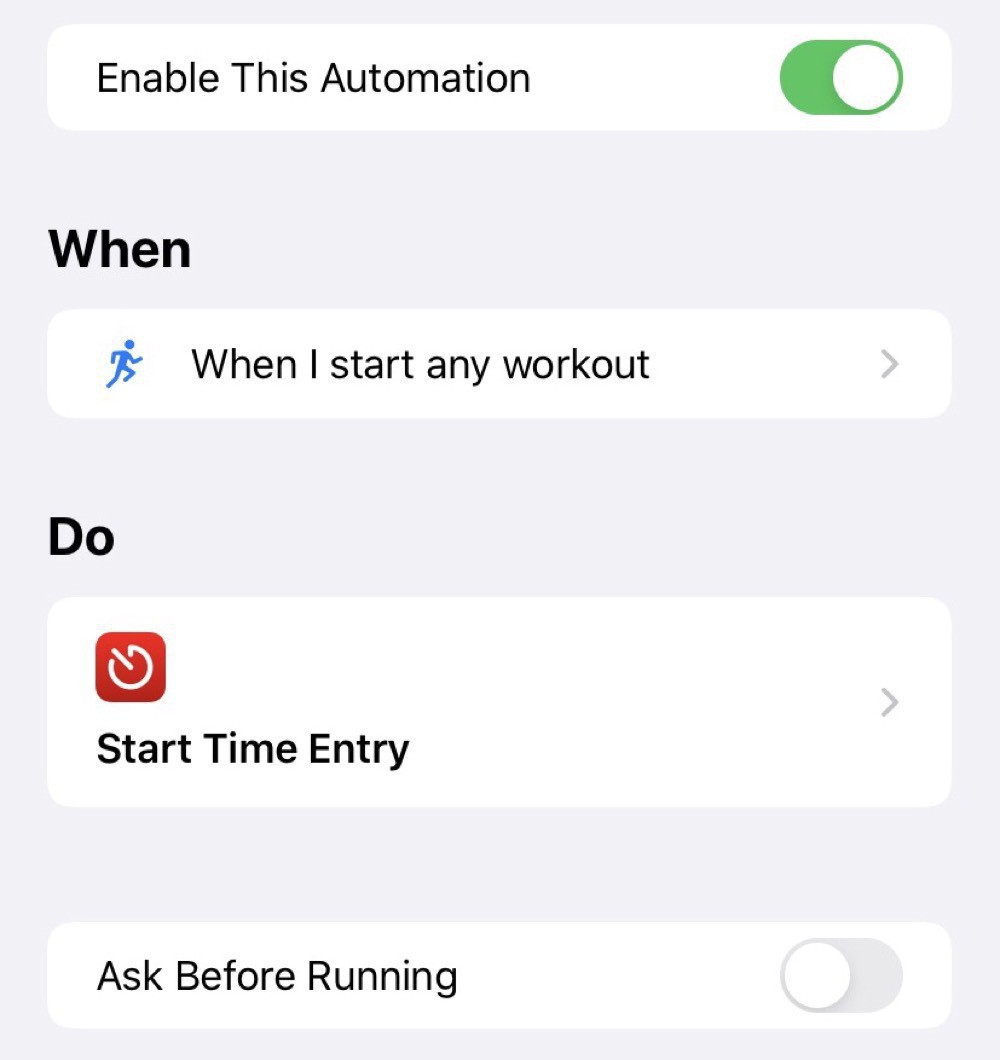
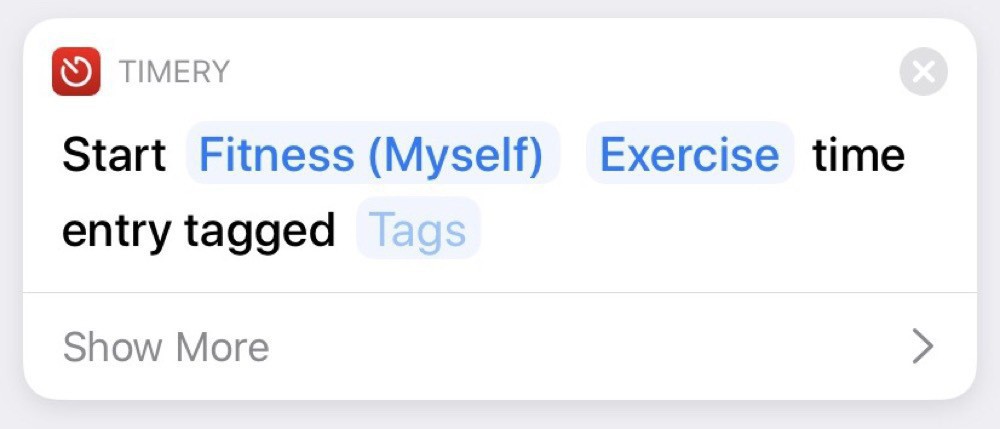
The starting automation just starts an entry in Timery:
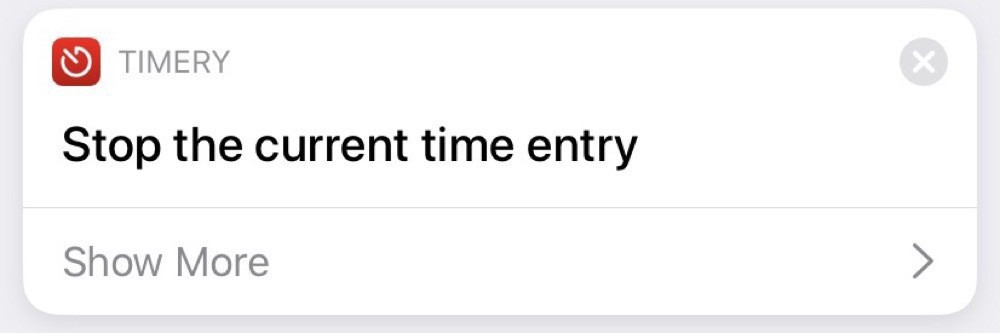
The stopping automation, unsurprisingly, stops the running entry:
As with most of my Shortcuts, this is a simple one. Developing a portfolio of these simple automations is really helpful for optimizing my processes and freeing up time for my priorities.
Brew coffee shortcut
Shorcuts in iOS is a great tool. Automating tasks significantly boosts productivity and some really impressive shortcuts have been created.
That said, it is often the smaller automations that add up over time to make a big difference. My most used one is also the simplest in my Shortcuts Library. I use it every morning when I make my coffee. All the shortcut does is set a timer for 60 seconds (my chosen brew time for the Aeropress) and logs 90mg of caffeine into the Health app.
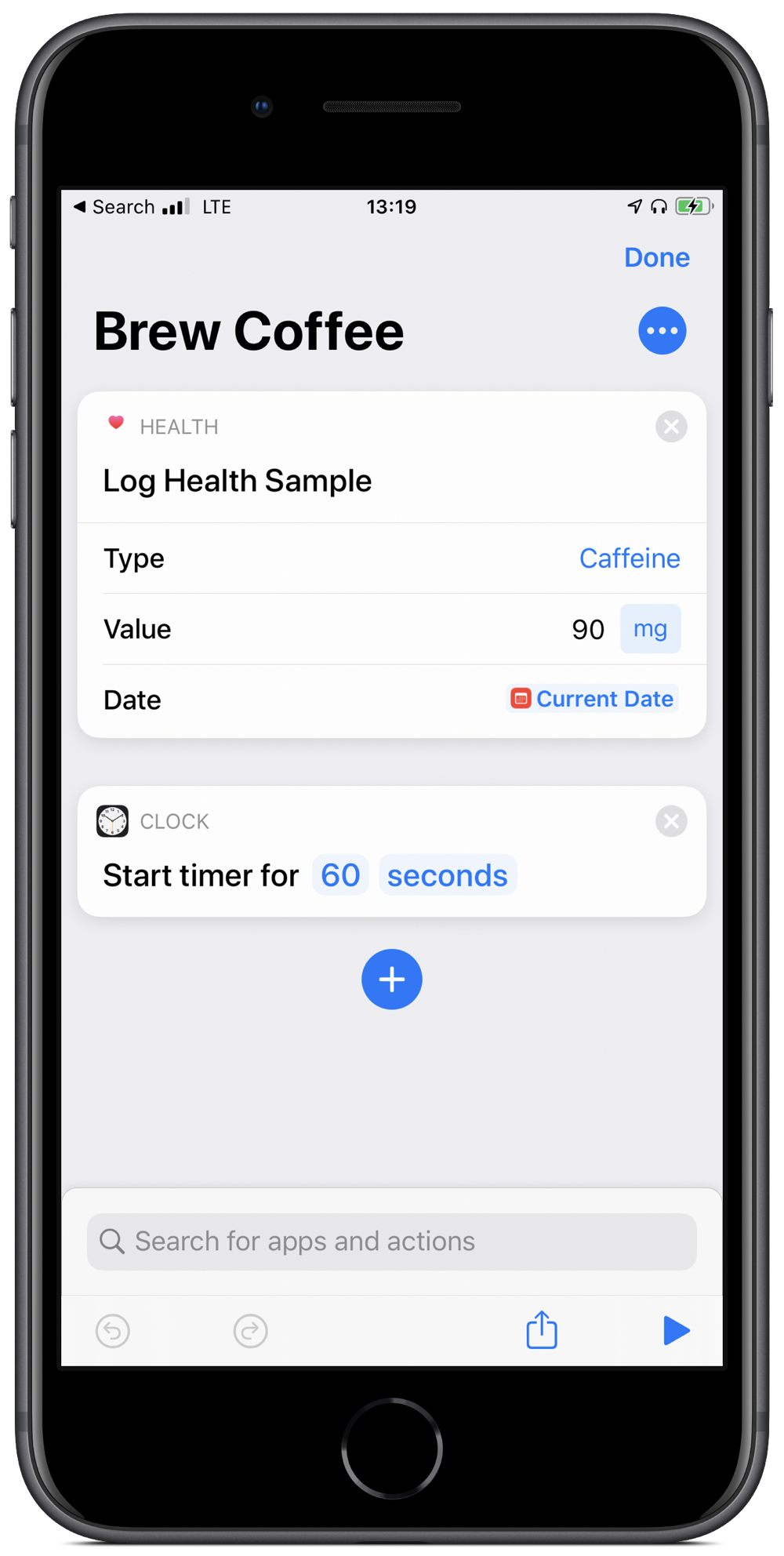
All I need to do is groggily say “Hey Siri, brew coffee” and then patiently wait for a minute. Well, that plus boil the water and grind the beans.
Simple, right? But that’s the point. Even simple tasks can be automated and yield consistencies and productivity gains.